A cellular traffic jam
UvA and VU researchers reveal jamming in cellular motor protein traffic
17 November 2017

Living cells require a constant transport of nutrition and waste. This is achieved by molecular motor proteins that transport organelles and other building blocks along the network of biopolymers of the cytoskeleton, which spans the volume of the cell. The walking mechanism of the individual motors has been studied extensively: Kinesin-1, for example, an important representative of the Kinesin family of proteins, moves by the subsequent, hand-over-hand stepping of two motor domains in well-defined steps of 8 nanometers.
Collective behaviour
What has so far remained unclear is how the motors walk and interact collectively. Due to their dense population, crowding effects could crucially affect the transport across the cell, but so far these effects could not be accessed in the densely populated regime. Doing so could be important, for example, in studying diseases like Alzheimer's disease, where it is well known that neuronal transport is severely hampered, resulting in local accumulations of motor proteins and their cargos, which could play a role in neurodegeneration.
Measuring velocity
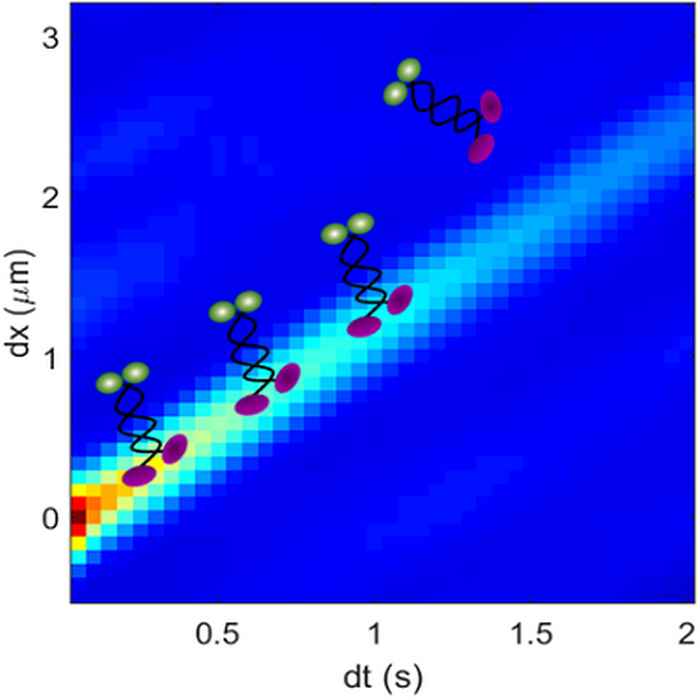
Researchers at the UvA and VU have now made significant progress on the jamming issue by combining a new correlation imaging technique with physical modelling. Like in previous studies, they used fluorescently labelled motors under well-defined conditions on microtubules - components of the cell's cytoskeleton - assembled on a glass slide. By correlating the moving image points of the fluorescent motor proteins in space and time, the researchers could for the first time measure their velocity and run length along the filament at high densities.
Traffic jams
These measurements revealed a remarkable slow-down of the motors as density increased, demonstrating the formation of traffic jams. These traffic jams were directly confirmed in the observed traces of the motors. Furthermore, the researchers showed that these traffic jams were well described by simple transport models, in which the motor proteins are modelled by hard particles that pile up as they get into each other’s way. Surprisingly however, the different motor species showed very different lengths over which they interact: from their physical size as assumed in the simple model, up to a distance 30 times larger than this size.
While clarifying the mechanism behind this long-range interaction remains an intriguing open problem for future research, the current results already illustrate the very different characteristics of the motors. Learning more about these motor protein-specific properties could help to cope with, or even suppress jamming effects in the cellular traffic.
Artist’s impression of molecular motors walking in space-time
Reference
Correlation Imaging Reveals Specific Crowding Dynamics of Kinesin Motor Proteins, D. M. Miedema, V. S. Kushwaha, D. V. Denisov, S. Acar, B. Nienhuis, E. J. G. Peterman, and P. Schall, Phys. Rev. X 7, 041037.
https://journals.aps.org/prx/abstract/10.1103/PhysRevX.7.041037